Automated CRISPR-Based Iterative Genome Engineering for Rapid Strain Devleopment & Improvement
Microbial strain improvement projects – such as protein or metabolic engineering – require many rounds of genome editing. The overall project timeline depends on the speed that you can move through the Design-Generate-Test-Learn (DGTL) cycle. The Onyx® Iterative Genome Engineering workflow streamlines the steps required to get from one genome engineering round to the next and allows you to move through the DGTL cycle much faster. To demonstrate what you can do with this workflow, we designed two applications:
- Heterologous protein engineering in E. coli and
- Improving the production of an industrially important recombinant enzyme in S. cerevisiae.
In both cases, the project took less than 6 months to complete.
For this strain development application, we engineered the heterologous green fluorescent protein (GFP) in the E. coli genome with a library of 723 targeted edits against known mutational hotspots. The first round of editing and screening identified variants with altered fluorescence characteristics, one of which showed a shift in the absorption-emission spectrum towards the yellow fluorescent protein (YFP). This strain was used to stack the same library of edits in the second round of genome engineering. As a result, two novel YFP variants were identified with significantly improved fluorescence.
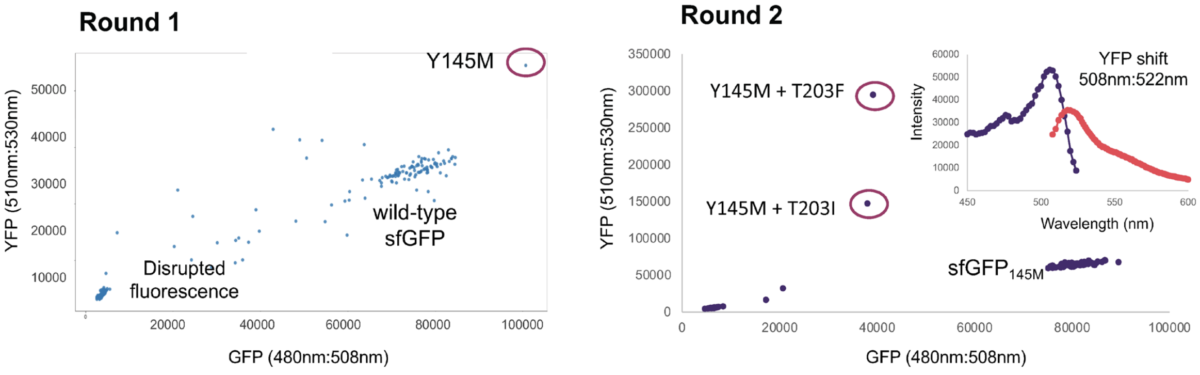
Discovery of two novel yellow fluorescent protein (YFP) variants through two cycles of Iterative Genome Engineering
The second application used a whole-genome approach to improve the production of heterologous cellobiohydrolase I (CBH1) enzyme in S. cerevisiae CEN.PK strain. We designed diverse libraries with >14,000 edits to target both known and unknown genes across many functional classes. After the first round of screening, we identified close to 100 hits producing up to 1.6‑fold improvement across different pathways, including transcriptional regulators, stress response, protein degradation, trafficking and glycosylation, and other genes.
To see if we can achieve further improvement by combining edits from different libraries, we took two top hits from the first round of screening and stacked a genome-wide knockout library on top of them. Additionally, we took the best performing strain from round 1 and stacked the remaining ~100 hits on top of it. These libraries showed up to 2.4‑fold improvement, and the best performing variant from round 2 was combined with the first-round hits for the third cycle of Iterative Genome Engineering. The final strain showed a ~3‑fold improvement in plate-based screening and a 4‑fold increase in CBH1 activity when tested in shake flasks.
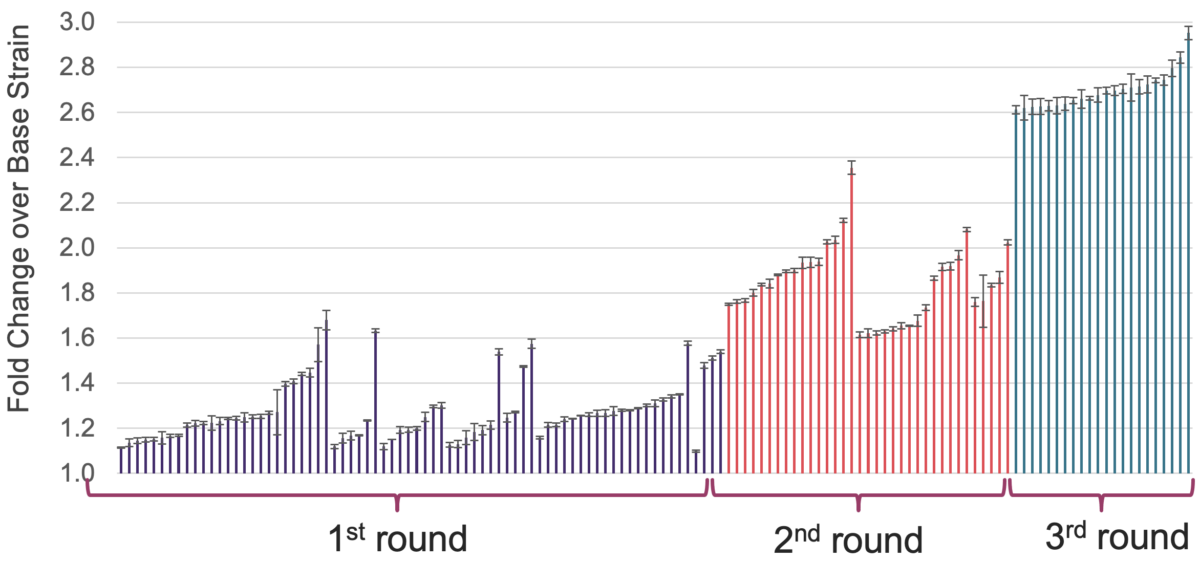
Fold-improvement in CBH1 activity over 3 cycles of Iterative Genome Engineering (plate-based screening)
Generate any library at the push of a button.
The Onyx gives you the flexibility and scalability to develop virtually limitless libraries. Create insertions, deletions, substitutions, or full saturation mutagenesis, in a single gene, a pathway, or the whole genome.
Resources & Support
Want to learn more about the Onxy platform and what we’ve been up to? Check out our latest videos, webinars, and presentations and get answers to our FAQs on the Resource & Support page.
- Copyright © 2025 Inscripta, Inc. All rights reserved.
- Legal Notices
- Privacy Policy
