Scalable Genome Engineering with push-button simplicity.
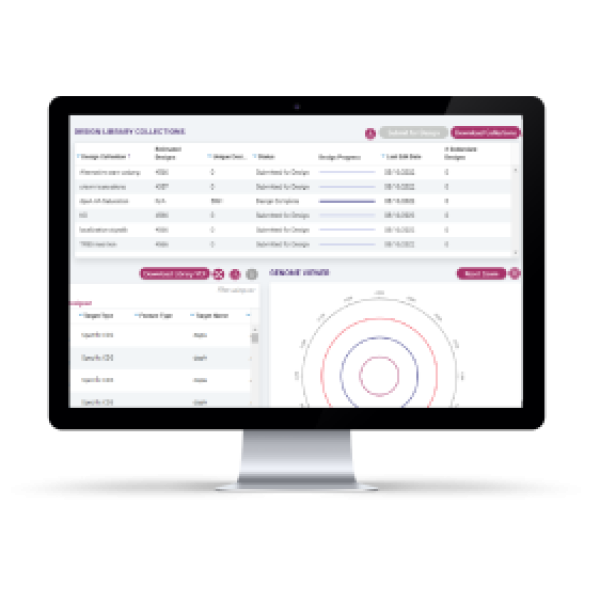
The Onyx® Platform is your answer for rapid microbial strain optimization. It’s a complete solution, consisting of design and analysis software, custom consumables, and a fully automated instrument. At the push of a button, you can design and generate complex libraries with thousands of edits, track results, and evaluate findings right on your benchtop.
Generate CRISPR-edited cell libraries with thousands of precise genomic edits in 2 – 4 days!