Facilitate your evolutionary studies
with the Onyx® platform
Adaptive laboratory evolution (ALE) approaches have long been a reliable way for researchers to discover functional mutations in cell populations cultivated for extended periods under well-defined stress conditions. ALE experiments are relatively inexpensive, simple to perform, and allow not only the discovery of adaptive mutations, but also the opportunity to study evolutionary routes by which those mutations are accrued.
While it can be a convenient exercise to recover and catalog a large and potentially interesting set of mutations using ALE approaches, not all changes are causal, and the deconvolution and validation side of the ALE program is often a time-consuming, laborious, and potentially costly affair.
We collaborated with one of our Early Access customers to apply Onyx technology to rapidly rebuild and assess a set of 372 previously collected ALE mutations in E. coli for which individual validation experiments had never previously been performed.
Additionally, because Onyx can do so much more, we expanded the library to test a few additional ideas by way of an extra 620 functional knockouts in the form of deletions, frameshifts, and mobile element insertions.
Where reconstruction and validation of a few hundred individual ALE mutations was once out of reach, a set of a thousand is now all within a day’s work using Onyx. By cultivating the entire library under conditions identical to the original ALE studies, we were able to quickly validate beneficial mutations by tracking enrichment of the associated barcodes within the population.
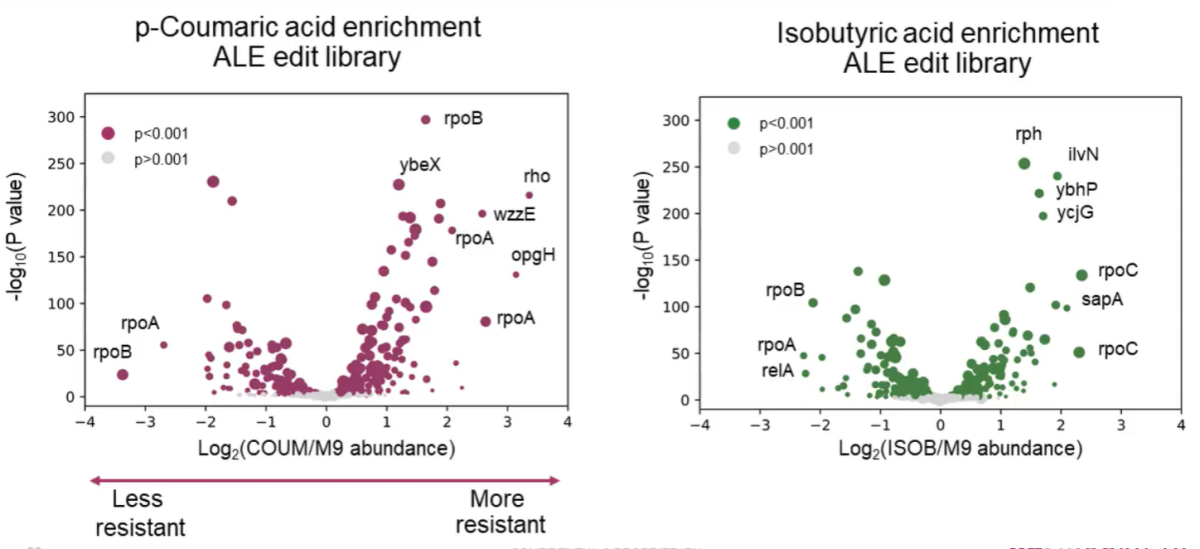
ALE study with Onyx cell libraries captures variants enriched or depleted under p‑coumatic and isobutyric acid pressure
Adaptive evolution mimics the processes that occur naturally in microbial populations, allowing us to study phenomena such as development of antibiotic resistance in bacteria. In another example of using the Onyx technology to facilitate ALE studies, our collaborators at the VIB-KU Leuven Center for Microbiology in Belgium were interested in elucidating the mechanisms behind bacterial persistence. The results were published in Molecular Biology and Evolution.
Bacterial populations were evolved and forced through varying bottleneck sizes. The Onyx technology was used to create a comprehensive pool of genetically diverse mutants from a library of 4,128 annotated loci and 87 ncRNA knockouts, which was then narrowed down by selection. Sequencing of evolved populations and knockout libraries after selection suggested different evolutionary routes and revealed genes involved in persistence, including previously known and novel targets.
Generate any library at the push of a button.
The Onyx gives you the flexibility and scalability to develop virtually limitless libraries. Create insertions, deletions, substitutions, or full saturation mutagenesis, in a single gene, a pathway, or the whole genome.
Resources & Support
Want to learn more about the Onxy platform and what we’ve been up to? Check out our latest videos, webinars, and presentations and get answers to our FAQs on the Resource & Support page.
- Copyright © 2025 Inscripta, Inc. All rights reserved.
- Legal Notices
- Privacy Policy
