Design. Discover. Repeat.
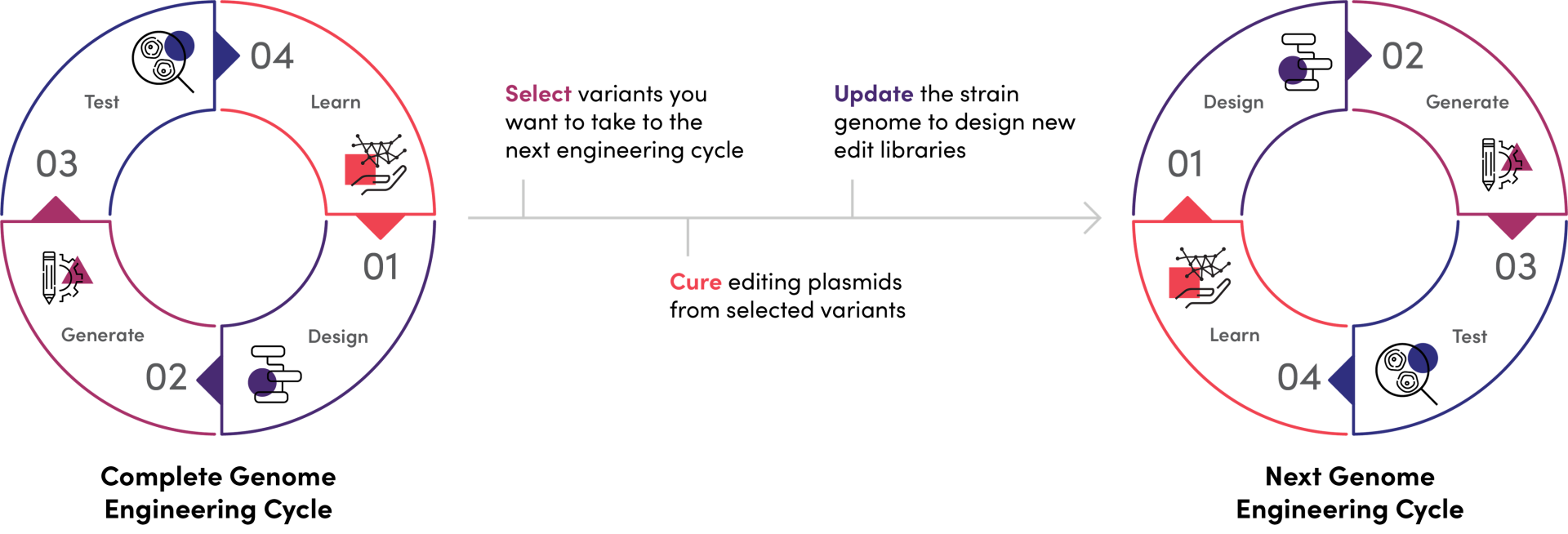
Iterative Genome Engineering allows you rapidly move through the Design-Generate-Test-Learn (DGTL) cycle to accelerate the discovery process. After screening your Onyx Cell Libraries and identifying the variants you want to take forward, you can design new edit libraries to generate additional diversity. To do that, the editing plasmids are cured from the engineered cells, which can be then transformed with new editing reagents. Update and import the new genome in just a few minutes using our software and proceed to design your next Genome Engineering Experiment.