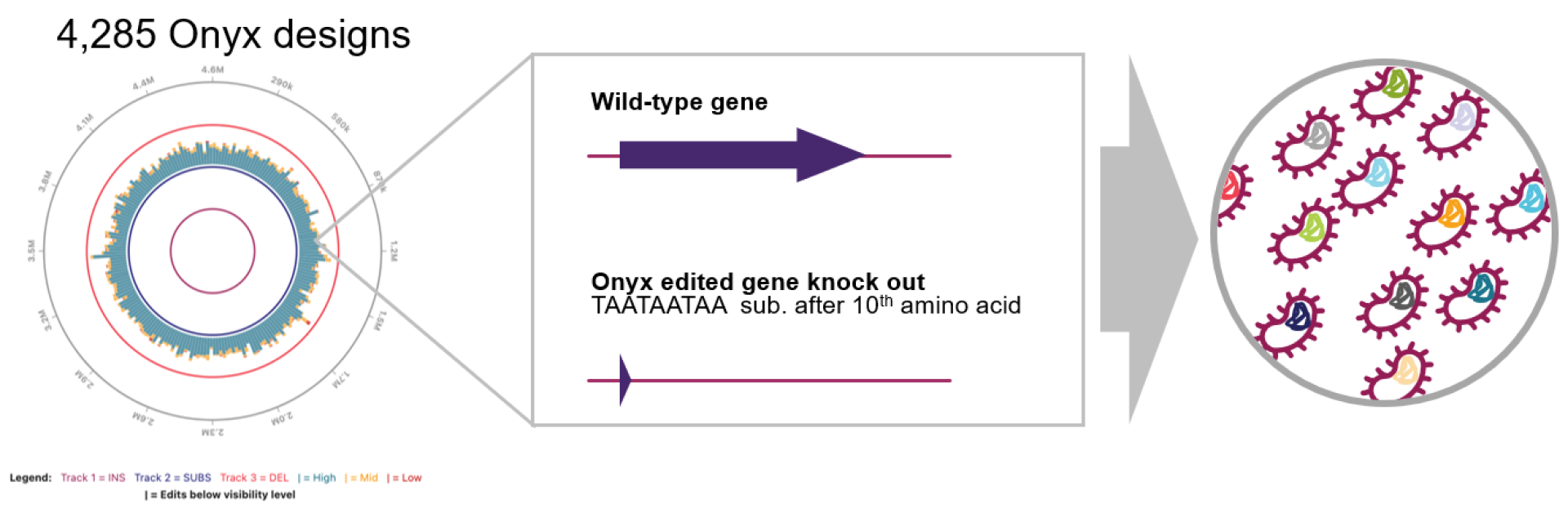
Your Favorite Library Series: Library 1: Genome-Wide Knockout (GWKO)
We’re kicking off a series of our favorite “library recipes” to showcase different library designs Inscripta scientists have created using the Onyx Platform®. This month we’re highlighting a knockout library strategy with each gene in the E. coli genome knocked out, created by Senior Scientist, Charlotte Cialek.
Why this is my favorite library:
The genome-wide libraries excite me because it allows for studying the entire genome in a single library pool.
Advantages of this library:
- Genome-wide libraries take genome discovery to the next level. There is still much to be discovered about gene function across microbial genomes (e.g. ~30% of the E. coli genome is termed the “y-ome” and still of unknown function1), so hits from genome-wide libraries can provide insights on the function of a previously unknown gene and can allow you to identify unexpected phenotypic improvements.
- Many gene knockouts have well-characterized phenotypes, which often enables a greater understanding of the underlying biology. I will often find myself going to look up any phenotypic hits from UniProt2, Ecocyc3, and other databases as soon as the InscriptaResolver™ come in!
- While the Keio collection has provided an invaluable resource for discovery in the E. coli BW25113 strain, imagine porting this level of discovery over to specific strains – that is one of the reasons this is often the first library I design when looking at a new strain.
- The genome-wide KO library is also great for dropping into screening and selection workflows. The barcode approach Onyx libraries utilize allows for tracking the behavior of individual edits within the entire population.
- One of my favorite approaches to take when looking for even more diverse variants is to take a top phenotypic hit and immediately cure and stack a GWKO library build on top of it. We often run into scenarios like our CBH1 experiment where this leads to much higher yields in just a few weeks!
Different approaches to create a GWKO library:
There are many different strategies for making a GWKO library, and the “best” strategy will depend on the intended purpose of the library. Below are some methods which can be used.
1. Stop codon insertion or substitution:
You can design 1–3 stop codons in frame or even in all three frames. We will often insert a triple stop codon sequence (TAATAATAA) after the 5th, 10th, or 15th amino acid in our knockout libraries, giving the ribosome a chance to fully initiate before termination at the chosen site. InscriptaDesigner™ software allows you to choose where you want to place your stop sequence in relation to amino acids (e.g. after the 10th amino acid), nucelotides, or as a relative fraction of the gene (e.g. substituting an every-frame stop codon TAATTAATTAA after 50% of each gene sequence for a genome-wide truncation library). You can get pretty creative with InscriptaDesigner, even substituting the nucleotides spanning the ribosome binding sites and start codons with a short hairpin sequence to remove translation initiation.
2. Transcription termination sequence:
Another approach may be to stop transcription altogether instead of only protein translation. Here you might use a short transcription terminator sequence (e.g., a T0 terminator) to substitute over nucleotides early in the operon of interest, effectively blocking downstream mRNA synthesis across the operon as well.
3. By design score:
The InscriptaDesigner software utilizes a powerful computational approach for optimal CRISPR-based editing, which includes a scoring system for each individual design. To optimize high design scores in your library, you can make a library with as many high design scores as possible for each gene. Using this strategy, I’ve generated and included the design recipe for our MG1655 strain here with the example of making a triple stop anywhere between the 5th amino acid to the 30th, with the highest binned design score being chosen. By doing a couple rounds of iterations and munging the libraries together, I was able to get the most possible designs with the highest binned design scores.
See what other libraries you can create and watch a video of the Designer software creating a KO library in a matter of minutes.
Download the CSV file for this KO library here.
Please note that specific designs for different organisms will vary. To discuss your specific library needs contact us.
References:
1. Ghatak, S., King, Z., Sastry, A., Palsson, B. (2019) The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function. Nucleic Acid Research, 47(5). https://doi.org/10.1093/nar/gkz030
2. Wang Y, Wang Q, Huang H, Huang W, Chen Y, McGarvey PB, Wu CH, Arighi CN (2021) A crowdsourcing open platform for literature curation in UniProt. PLoS Biol. 19(6). https://doi.org/10.1371/journal.pbio.3001464
3. Keseler, I., Collado-Vides, J. et. al. (2011) EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acid Research 39. https://doi.org/10.1093/nar/gkq1143